舞茸蛋白多糖通过激活BAK-1基因诱导乳腺癌细胞凋
日期:2019-07-31 / 人气:
声明:本文转载自Journal of Medicinal Food ,即《药物强化性食品杂志》,是由美国Mary Ann Liebert出版公司出版发行的一本有关功能食品及保健食品的国际性杂志。为方便广大读者阅读,我们对论文摘要进行了中文翻译,译文仅供参考,如有不足之处,敬请指正。
【参考译文】
Maitake (D Fraction) Mushroom Extract Induces Apoptosis in
Breast Cancer Cells by BAK-1 Gene Activation
舞茸蛋白多糖通过激活BAK-1基因诱导乳腺癌细胞凋亡
摘要:根据多年经验,菇类一直被用作传统药物治疗多种疾病。基于对舞茸固有的免疫调节和抗肿瘤特性的研究,人们进一步分离出多种具有生物活性的复合物,其中之一的舞茸蛋白多糖已被确认具有削弱肿瘤细胞生存能力的功能。本研究验证了舞茸蛋白多糖对人类乳腺癌细胞(MCF7)生存能力和细胞凋亡的影响。这些细胞分别用浓度为18μg/ml, 36μg/mL, 91μg/m, 183μg/mL, 367μg/mL的舞茸蛋白多糖进行处理,另有一组未经处理的细胞作为对照组,实验持续24小时。根据舞茸蛋白多糖的剂量,处理过的细胞生存能力(根据3-(4,5-二甲基)-5-(3-羧甲基苯环)-2-(4-硫基苯)-2H-四唑盐复合物检测法)依次有所降低。细胞凋亡的统计数依剂量都有明显上升(终端脱氧核糖核酸末端转移酶介导的三磷酸脱氧尿嘧啶缺口端标记法)。细胞经蛋白多糖恒温培养后,进行微阵列检测,结果显示能直接体现细胞凋亡途径的两种蛋白质——BAK-1基因和细胞色素C的转录产物上升。反转录聚合酶链反应也证实了微阵列检查的结果:BAK-1是最过表达的基加盟“|6因之一。这诸多发现更加肯定了舞茸蛋白多糖对乳腺癌细胞凋亡的影响,并进一步凸显了释放于细胞质的细胞色素C的干预作用。释放在细胞质中的细胞色素C,是细胞凋亡途径中另一个主要因素,其产量在经舞茸蛋白多糖培养后有所增加,且随蛋白多糖浓度的增加而上升。这一发现显示,舞茸蛋白多糖的作用在于诱导线粒体功能障碍。因此,阐明舞茸蛋白多糖发挥作用的分子机理对于癌症预防和治疗方法的发展至关重要。
【论文原文】
Journal of Medicinal Food
2011, 1-10
Maitake (D Fraction) Mushroom Extract Induces Apoptosis
in Breast Cancer Cells by BAK-1 Gene Activation
Raquel Soares, Manuela Meireles,1 Ana Rocha,1 Ana Pirraco,1 Diego Obiol,2 Eliana Alonso,2
Gisela Joos,2 and Gabriela Balogh2
1Department of Biochemistry, Faculty of Medicine, University of Porto Foundation, Porto, Portugal.
2Center for Scientific and Technical Investigation, Cerzos-Conicet, Bahıá Blanca, Argentina.
ABSTRACT For many years mushrooms have been used empirically in traditional medicine to treat several diseases. Study of the maitake mushroom, with its immunomodulatory and antitumoral properties, has led to the isolation of several bioactive compounds. One of these, D fraction, is known to reduce tumor cell viability. This study examined the effect of isolated D fraction on viability and apoptosis of human breast cancer cells (MCF7). These cells were treated with maitake (D fraction) extract at 18μg/mL, 36μg/mL, 91μg/mL, 183μg/mL, or 367μg/mL or were left untreated (control) for 24 hours. MCF7 incubation with the maitake extract resulted in decreased cell viability [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophe- nyl)-2H-tetrazolium assay] in a dose-dependent manner. Apoptosis was statistically significantly increased in a dose-dependent manner at every concentration tested (terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick-end labeling assay). Upon incubation with D fraction, a microarray assay revealed upregulation of BAK-1 and cytochrome c transcripts, 2 proteins directly involved in the apoptotic pathway. Reverse transcriptase polymerase chain reaction studies confirmed these findings; BAK-1 was one of most overexpressed gene, as observed by microarray assay. These findings confirm the apoptotic effect of maitake D fraction in breast cancer cells and further highlight the involvement of cytochrome c release to the cytoplasm. Cytoplasmic release of cytochrome c, another player in the apoptotic pathway, was also increased after incubation with D fraction in a dose-dependent manner. This finding indicates that the effect of this compound involves mitochondrial dysfunction. The identification of the molecular mechanisms by which D fraction exerts its effects is crucial for the development of preventive and therapeutic strategies for cancer
.
KEY WORDS: ·apoptosis·BAK-1·breast cancer·cytochrome c·maitake·mushroom
INTRODUCTION
Breast cancer is the leading cause of death from cancer in women, the most common cancer among women worldwide, and the second most frequent cancer in the world. Its incidence is still increasing, with a total number of cases of 1,050,100 in 2002 compared with 572,100 in 1980. Breast cancer causes 370,000 deaths annually, representing 13.9% of cancer deaths in women, mainly in the industrialized countries.1 While researchers search for the cause of the disease, an effective and rapid treatment is mandatory for these patients.
Maitake mushroom (Grifola frondosa) is a giant mushroom indigenous to northern Japan that has been used by Japanese herbalists for many years. Most research on maitake has focused on the use of maitake D fraction for treating various malignancies. The bioactive D fraction extracted from maitake is a protein-bound polysaccharide compound and is prepared by a standardized procedure developed by Maitake Products Inc. Among the various maitake fractions that have been prepared, D fraction was found to be the most potent for enhancing the immune system via oral administration or injection (both routes were effective), leading to the highest reduction in the rate of cancer proliferation.2 Maitake D fraction has been reported to exert its antitumor effect in tumor-bearing mice by enhancing the immune system through activation of macrophages, T cells, and natural killer cells.3 In a previous study, the combination of immunotherapy with the maitake D fraction and chemotherapy suggested that the D fraction might decrease the size of lung, liver, and breast tumors in patients with cancer.2,4,5 In 1998, the Food and Drug Administration granted Maitake Products, Inc., an investigational new drug application to conduct a phase II pilot study using maitake D fraction among patients with advanced breast and prostate cancers. These ongoing studies are evaluating the immune stimulatory effect of D fraction on tumor size, immune assays, clinical symptoms, and patient quality of life.
The purpose of the current study was to investigate the direct effect of maitake D fraction on breast cancer MCF7 cells. We found that incubating MCF7 cells with maitake D fraction significantly decreased cell viability and induced cell death. Genomic analysis using complementary DNA (cDNA) microarrays revealed the upregulation of 22 pro-apoptotic and 7 anti-apoptotic genes compared with untreated MCF7 cells (control). BAK-1 transcript at the messenger RNA level was found to be upregulated. After incubation with D fraction, cytoplasmic release of cytochrome c, another player in the apoptotic pathway, was also increased in a dose-dependent manner.
MATERIALS AND METHODS
Bioactive maitake D fraction
The bioactive D fraction was extracted from maitake mushroom, corresponding to the protein-bound polysaccharide compound, and was prepared by a standardized procedure developed by Maitake Products, Inc.
Cell cultures
The human breast cancer MCF7 cell line was obtained from the American Type Culture Collection. MCF7 cells were routinely cultured in Eagle minimal essential medium containing 10% inactivated fetal bovine serum and 1% penicillin/streptomycin. Cell culture media, fetal bovine serum, and penicillin/streptomycin were purchased from Invitrogen Life Technologies. Cells were grown at 37℃ in a humidified 5% CO2 atmosphere. In agreement with previous studies, incubations were performed for 24 hours in serum-free conditions.6
Labeling and cDNA human microarrays
We adopted the direct labeling of probes with amine-modified random primer, using 5μg of amplified RNA (aaRNA) as starting material. The 5μg of aaRNA (8μL) was combined with amine-modified random primer (2μg/μL,1μL) and ribonuclease inhibitor (5 units/μL, 1μL). The mix was incubated at 70℃ for 10 minutes and then chilled on ice for 10 minutes. Primer-RNA solution was added to the reverse transcriptase mix (5×first-strand buffer, 6μL; 50× amplified deoxyuridine triphosphate [dUTP]/dinucleotide triphosphate [25mM deoxyadenosine triphosphate, deoxyguanosine triphosphate, and deoxycytidine triphosphate; 15mM deoxythymidine triphosphate; and 10mM aminoallyl-dUTP], 0.6μL; dithiothreitol, 0.1M, 3μL; Superscript II reverse transcriptase [Invitrogen/Life Technologies], 2μL) and incubated at 42℃ for 2 hours. The reaction was terminated by adding EDTA (0.5M, 10μL), and the RNA was hydrolyzed with NaOH (1M, 10μL) at 65℃ for 30 minutes.
Probe purification
Probes were cleaned with a QIA quick polymerase chain reaction (PCR) purification kit (Qiagen); the Cy3- and Cy5- labeled products were combined and 30μL of water was added, followed by 500μL of Buffer PB. The samples were applied to QIA quick columns, which were centrifuged at 13,000 rpm for 1 minute, after which the flow through were discarded. To wash the columns, 750μL of Buffer PE was added, columns were spun again for 1 minute, and the flow-through was discarded. The washing step was repeated once more, and columns were spun again to remove residual ethanol. Fresh collection tubes were placed beneath each column, 30μL of Buffer EB was added, and tubes were incubated for 1 minute at room temperature. Columns were then centrifuged at 13,000 rpm for 1 minute, and the elution step was repeated once. Elutes were partially dried in a vacuum centrifuge and the volumes were adjusted to 23μL with water.
Hybridization and washing conditions
The following were added: 4.5μL of 20×saline-sodium citrate, 2μL of poly(A) (10mg/mL), and 0.6μL of 10% (wt/vol) sodium dodecyl sulfate (SDS). The probes were then denatured at 98℃ for 3 minutes. The products were pipetted onto arrays, coverslips were applied, and the slides
were placed in a hybridization chamber (Corning). cDNA human microarrays containing 25,000 known genes were incubated in a 42℃ water bath for 16 hours and were subsequently washed with 0.5×saline-sodium citrate, 0.01% (wt/vol) SDS, followed by 0.06×SSC, at room temperature
for 10 minutes each. Slides were spun for 5 minutes at 800rpm (130 g) at room temperature.
Array scanning and analysis
Arrays were read with an Affymetrix 428 fluorescent scanner (MWG Technologies) at 10-μm resolution and variable photomultiplier tube voltage settings to obtain the maximal signal intensities with less than 1% (wt/vol) probe saturation. The resulting images were analyzed by using ImaGene, version 4.2, and GeneSight software, version 4.0 (Biodiscovery, Inc.). After Lowess normalization, the data were analyzed by using GeneSigh 4.0 software (Biodiscovery).
Hierarchical clustering analysis
Hierarchical clustering was done by using the neighbor-joining method applied to the Pearson correlation distance matrix. In addition, other hierarchical clustering methods and other distance-generating functions (such as Euclidean) were used. Only marginal deviations between the methods and the distance matrices used were observed. We found the most stable results for the neighbor-joining method applied to the Pearson correlation distance matrix.
MTS toxicity assay
MCF7 cells (2×105 cells/mL) were allowed to grow until 70-90% confluence and were incubated with each treatment for 24 hours. Cells were washed twice with phosphate-buffered saline, and their viability was assessed by using Cell Titer 96 Aqueous ONE Solution Reagent (MTS [3- (4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H- tetrazolium] colorimetric assay (Pro-mega), according to manufacturer’s instructions. Optical density was measured at 492nm. Results are expressed as percentage of control, which was considered to be 100%.
TUNEL assays
Cells grown until 70% confluence onto glass coverslips were incubated with the different referred treatments for 24 hours. A TUNEL assay (terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick-end labeling) was performed by using the In Situ Cell Death Detection kit (Roche Diagnostics), according to the manufacturer’s instructions. Nuclei were counterstained with 4-6-diamidino-2-phenylindole (DAPI) (Roche Diagnostics). The percentage of TUNEL-stained nuclei was evaluated in relation to every DAPI-stained nucleus observed. Immunofluorescence was visualized under a fluorescence microscope (Olympus, BH-2). The percentage of stained cells was evaluated by counting the cells stained with TUNEL divided by the total number of nuclei stained with DAPI at×200 magnification field. One thousand nuclei were evaluated. Three independent experiments were performed.
Reverse transcriptase PCR
Reverse transcriptase (RT) PCR was performed to confirm the results obtained by cDNA array hybridization in MCF7 cell lysates. For each sample, 1.0μg of RNA was reverse-transcribed in a reaction volume of 25μL in the presence of 10mM dNTP and 2μL of RT-PCR enzyme mix. Gene-specific primers for BAK-1 (forward: GAGAGCCTGCCCTGCCCTCT; reverse: CCACCCAGCCACCCCTCTGT), coamplified with a set of primers for the house-keeping gene βActin, were used in RT-PCR reaction. Quantifications were done in triplicate. The PCR products
were separated on ethidium bromide-stained 2% agarose gel. The intensity of the fluorescence was automatically measured and integrated by using Genescan software (Image Master, Pharmacia).
Western blotting
Proteins were isolated from MCF7 total cell lysates by using Tripure (Roche Diagnostics). For cytoplasmic protein isolation, MCF7 cells cultures were subjected to the Abcam cell fraction protocol (Tufts-New England Medical Center and Molecular Cardiology Research Center). In brief, cells were lysed with 500μL of cell-fractioning final solution and kept on ice for 20 minutes. Protein cytoplasmic fraction was then obtained upon centrifugation at 8000 rpm for 10 minutes. Pellets were then washed and proteins were quantified by using the bicinchoninic acid protein quantification assay kit (Pierce Biotechnology). Equal amounts of protein were subjected to 10% SDS-polyacrylamide gel electrophoresis with a 5% stacking gel. After electrophoresis, proteins were blotted into a Hybond nitrocellulose membrane (Amersham Biosciences) by using a mini-transblot electrophoretic transfer cell (Amersham Biosciences). Immunodetection for cytochrome c and βactin (both at 1:2000 dilution) (Santa Cruz Biotechnology) was accomplished with enhanced chemiluminescence (ECL kit, Amersham Biosciences). Anti-goat secondary antibody conjugated with horseradish peroxidase (1:5000) was obtained from Santa Cruz Biotechnology. The relative intensity of each protein blotting analysis was measured by using a computerized software program (Biorad) and normalized with βactin bands to compare the expression of proteins in different treatment groups. Experiments were repeated 3 times.
Statistical analyses
All experiments were performed in triplicate. Quantifications are expressed as means±standard deviations. Samples were evaluated by an analysis of variance test. Differences between experimental groups were analyzed by the Student t-test and were considered statistically significant whenever the P value was less than .05. Statistical analysis was performed using GraphPad.
RESULTS
Maitake induces the expression of apoptotic genes in breast cancer cells
Our objective was to determine whether D fraction from G frondosa can induce the apoptotic pathway in MCF7cells. We performed a cDNA microarray using 25,000 known human genes. MCF7 cells were treated with increased concentrations of D fraction over 24 hours. Figure 1A shows a representative cDNA microarray image from each of the MCF7 cells treated with and without increased concentrations of maitake D fraction. In the right portion of Figure 1A, “a” corresponds to the square section indicated in the left microarray image. Three independent experiments were done. Figure 1B represents the box plot analysis after Lowess normalization under all conditions. Genes whose expression changed were considered statistically significant according to established algorithms, and the genes whose expression changed by at least 2-fold as a result of Maitake treatment were selected for further analysis. This combined analytic approach has previously been shown to identify differentially expressed genes with high sensitivity and specificity.
We identified 4321 gene sequences differentially expressed (t-test with false discovery rate, P<.05) after incubation with D fraction (at 91μg/mL or 187μg/mL or 367μg/mL during 24 hours) on MCF7 cells. Unsupervised hierarchical clustering performed by using the expression profiles of 4321 globally varying genes across the controls and Maitake-treated MCF-7 cells (data sets representing the 4 groups) revealed that samples clustered primarily according to D fraction treatment. Figure 1C represents the unsupervised hierarchical cluster 3 in all conditions. This finding suggested that the principal source of global variation in gene expression across these data sets was due to increased Maitake concentrations. In Figure 1C, black arrows indicate the specific variation in gene expression related to apoptosis due to Maitake treatment.

FIG. 1. Analysis of gene expression induced by maitake in breast cancer MCF7 cells. (A) Images of the cDNA microarrays employing 25,000 human known genes from MCF7 treated with and without (control) increased concentrations of maitake D fraction. The magnified right images (a) correspond to the indicated white square in each condition. The cDNA microarrays were performed against the human reference and used Cy3 fluorescent dye. In the microarrays, the green spots correspond with the Cy3 channel or the human reference; in the red channel (Cy5 dye) with red spots, we observed the genes overexpressed in relation to the control and the MCF7 treated cells. The yellow spots indicate the genes that were expressed in both channels (that is, were expressed equally for the human reference and for the sample). (B) The box plot analysis after Lowess data normalization, performed for each condition (see Materials and Methods section). (C) Two-dimensional unsupervised hierarchical clustering of the data matrix, consisting of 25,000 genes by 4 samples and representing the control and maitake-treated MCF-7 cells. Rows represent genes and columns represent sample conditions. The cDNA microarray experiments were performed in triplicate. Black arrows (c) indicate the 29 apoptosis related-genes that were significantly overexpressed after maitake treatment (Table 1).
Only 29 apoptotic genes were statistically significantly upregulated after maitake treatment (Table 1). BAK-1 was one of the pro-apoptotic genes with the most increased expression after incubation with D fraction. Accordingly, expression of BAK-1 was upregulated by D fraction treatment in a dose-dependent manner; expression was increased almost 25-fold upon incubation with 367μg/mL of D fraction when compared with controls (Table 1). Table 1 lists these 29 genes and the average ratio value with respect to control. Four of 29 genes were related to induction of apoptosis (pro-apoptotic genes): BAK-1 (BCL2-antagonist/killer 1), BCL2L13 (BCL2-like 13 [apoptosis facilitator]), BID (BH3 interacting domain death agonist), and NUPR1 (nuclear protein 1). One gene, cytochrome c1 (CYC1), was related to electron transport from mitochondria; 17 were directly involved in apoptosis; and 7, including MPO (myeloperoxidase), TNFAIP8 (tumor necrosis factor α-induced protein 8), VDAC-2 (voltage-dependent anion channel 2), and BCL2L2 (BCL2-like 2), were related to anti-apoptosis (anti-apoptotic genes). We decided to begin studying the apoptotic pathway with one of the genes most activated by maitake in the MCF7 cells.
Maitake directly induces cells death in breast cancer cells
To determine whether maitake D fraction can directly induce cell death in MCF7 cells, we treated cells with increased concentrations of maitake and evaluated cell viability and cell death by using a Time Lapse Microscope; pictures were taken every 10 minutes for 24 hours. Figure 2A shows a representative image (from 3 independent experiments) of MCF7 cell culturing treated with and without maitake at different concentrations. These correspond to the images taken 1 hour, 5 hours, 10 hours, and 24 hours after treatment. Microscope images were set up at×20 amplification. After we analyzed the video data and counted the relative percentage of cell deaths under each condition, we found a significant increase in cell death with dose-dependent maitake treatment; apoptosis was highest at the maximum concentration of 367μg/mL, reaching almost 98% cell death (Fig. 2B). We observed a high percentage of cell death under all conditions because of the stress (incubation at room temperature and not-humidified atmosphere controlled) that cells suffer with the Time Lapse equipment. The atmosphere was not controlled for humidity.
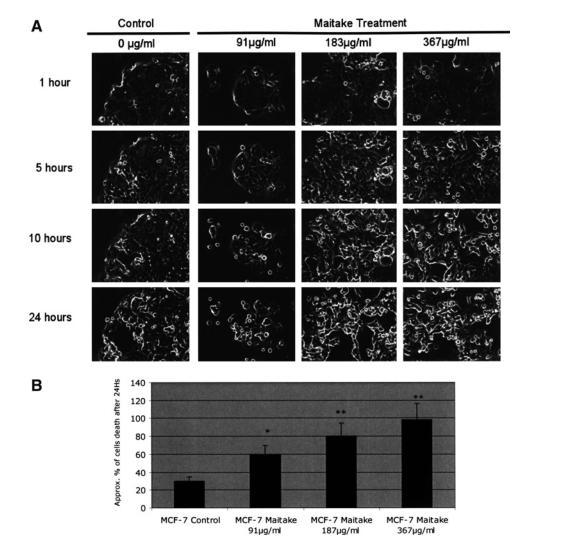
FIG. 2. Analysis of cell death induced by maitake in MCF7 cells using the Time Lapse microscope. MCF7 cells at 70% confluence were treated with and without (control) increased concentrations of maitake D fraction. The experiments were performed in triplicate. Cells were placed in the Time Lapse microscope under a CO2 atmosphere at room temperature for 24 hours. The camera was set up to take pictures every 10 minutes, using the×20 objective. Images and video were analyzed by using specific software. (A) The representative image corresponding to the cell culture at 1 hour, 5 hours, 10 hours, and 24 hours after maitake treatment in the conditions indicated in the figure. (B) The approximate percentage of dead cells observed in each image after 24 hours of maitake incubation.
D fraction decreased MCF7 cell viability and increased apoptosis
The evidence that maitake D fraction exerted anticancer effects and directly induced cell death prompted us to measure the effect of this compound on viability of MCF7 cells and to examine whether the trigger mechanisms for cell death are related to apoptosis. Cell viability was first examined with MTS assay in MCF7 cell cultures incubated with 5 different concentrations of D fraction. The number of viable cells decreased gradually with increasing concentrations of D fraction (Fig. 3). In fact, the highest concentration of D fraction resulted in a significant decrease in viability compared with control values (Fig. 3).
To evaluate whether this decrease in cell viability was due to increased apoptosis, a TUNEL assay was performed. MCF7 (4×104) cells were incubated with the same 5 concentrations of maitake D fraction for 24 hours, and the percentage of apoptotic cells was quantified. Treatment with this fraction, at any concentration, led to a statistically significant increase in the number of apoptotic cells in a dose-dependent manner (Fig. 3B). Of note, nearly 95% of the cells became apoptotic when treated with the highest concentration of maitake fraction (367μg/mL) (Fig. 3B). Figure 3C shows a representative image of these latter findings. As observed, treatment with this fraction increased the number of apoptotic cells (green) compared with the untreated culture.
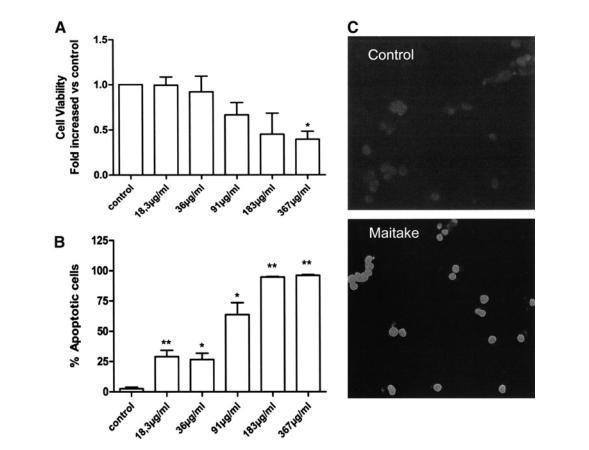
FIG.3. MCF7 cell viability was evaluated after incubation with 5 concentrations of maitake D fraction. (A) Cell viability was assessed by MTS assay. Results are expressed in absorbance values at 540nm and are the fold-increase relative to control cell cultures. Three independent experiments were performed in triplicate, with identical results. *P=.05 vs. control (n=9). (B) Apoptosis was increased at every incubation as evaluated by TUNEL assay, reaching statistical significance for every concentration of D fraction. *P<.05 vs. controls; **P<.01 vs. Control. Bars represent the percentage of apoptotic cells evaluated by the ratio between TUNEL-stained cells and DAPI-stained nuclei in every culture. Experiments were performed in triplicate, with identical results (n=9). (C) Immunofluorescence for apoptotic cells (green). Note the increased number of apoptotic cells in cells treated with 367μg of maitake fraction D (bottom panel) in comparison to untreated cells (top panel). Representative images are shown. Magnification×200.

Maitake increased BAK-1 gene expression in MCF7 cells
Expression of the BAK-1 gene was greatly increased after incubation with D fraction. Accordingly, expression of BAK-1 was upregulated by D fraction treatment in a dose-dependent manner. BAK-1 transcript was examined by using RT-PCR assay. As illustrated in Figure 4, the messenger RNA level of BAK-1 increased upon treatment with maitake D fraction at increasing concentrations, confirming the previous findings obtained by microarray assay. In these assays a house keeping gene, βactin, was used as a control for gene expression. Figure 4 shows the ratio of BAK-1 gene expression with respect to βactin (top panel) and the RT-PCR-specific bands at increased maitake concentrations (bottom panel).
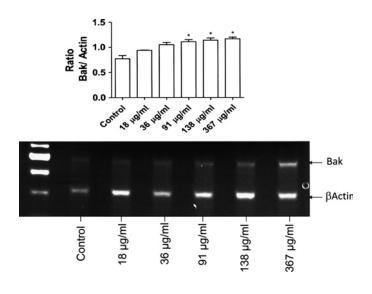
FIG. 4. Expression of BAK-1 in MCF7 whole cell lysates after incubation with different concentrations of maitake D fraction over 24 hours. Reverse-transcriptase polymerase chain reaction analysis for BAK-1 and βActin was carried out by using RNA derived from vehicle-treated (control) and 5 concentrations of maitake D fraction MCF7 cell cultures. BAK-1 was upregulated by the compound in a dose-dependent manner. Bars correspond to intensity ratios of BAK-1 gene expression after normalization to βActin. Results are representative of 3 independent experiments. Statistically significant differences were found for incubation with the 3 highest D fraction concentrations. *P<.05 vs. controls.
Maitake increased cytochrome C cytoplasmic level in MCF7 cells
To further investigate the possible mechanisms by which maitake D fraction exerts its apoptotic effects, the expression of cytochrome c was addressed. Cytoplasmic lysates were obtained from cultures previously incubated with the 5 different concentrations of maitake D fraction or were left untreated for 24 hours; cytochrome c expression was then examined by Western blotting. Although no significant findings were observed, a dose-dependent increase in the cytoplasmic cytochrome c occurred upon treatment with the compound. This finding indicates that D fraction promoted cytochrome c release from the mitochondria (Fig. 5).
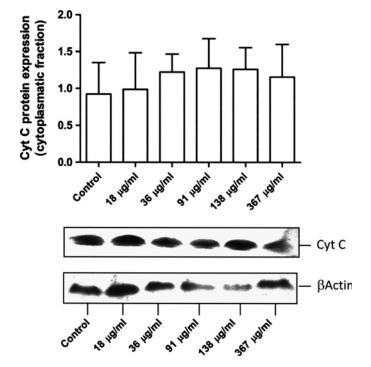
FIG.5. Immunoblotting for cytochrome c after incubation with maitake D fraction. Increased expression of cytochrome c was observed after incubation with increasing doses of D fraction, although this finding was not statistically significant. Equivalent protein loading was confirmed by probing stripped blots for βActin, as shown. The bottom image shows a representative Western blot sample from 3 independent experiments (n=6). Cyt c, cytochrome C.
DISCUSSION
Breast cancer mortality has remained almost unchanged for the past 5 decades. The condition is second to lung cancer as a cancer-related cause of death.7,8 The failure to eradicate this disease is largely due to the inability to identify a specific etiologic agent, determine the precise time of initiation, and resolve the molecular mechanisms responsible for cancer initiation and progression. Until the exact cause of breast cancer is found, researchers are investigating antitumoral agents and their potential clinical applications. An effective therapy is becoming increasingly important in fighting this disease. Current breast cancer treatments involve surgery, followed by radiation and hormonal therapy or chemotherapy. Some of these treatments have toxic side effects. The drugs used in chemotherapy affect normal cells as well as cancer cells. Researchers are trying to develop targeted drugs that attack only cancer cells and do not harm normal cells. One approach is to look for compounds that can block genes involved in transforming a normal cell into a cancer cell.
Recently, several in vitro studies have shown that pentamer (from the family of flavonoids derived from chocolate) inhibits growth of breast cancer cells.9,10 In addition, for thousands of years, humans have used medicinal mushrooms because of their healing potential. A rich folkloric tradition has passed the knowledge of mushrooms down through the ages, particularly in Japan and China, where mushrooms are supported by decades of scientific research and are imbedded in the cultural consciousness. U.S. researchers have been exploring these fascinating organisms for their medicinal properties. The future for mushrooms as a nutritional supplement looks bright as long as clinical studies continue to support the mushroom’s properties as a powerful healing agent with the ability to improve human health.
In this study we demonstrated that D fraction of the maitake mushroom induced apoptosis in MCF7 breast cancer cells. These findings were corroborated by a reduction in cell viability with 5 different concentrations of this fraction. Although several genes promoting apoptosis were upregulated, as assessed by cDNA microarray analyses, the current study examined BAK-1 and cytochrome c within these cells. Our RT-PCR BAK-1 results confirmed the increased expression of BAK-1. The BAK-1protein localizes to mitochondria and induces apoptosis by accelerating the opening of the mitochondrial voltage-dependent anion channel (VDAC), which leads to a loss in membrane potential and the release of cytochrome c. Pro-apoptotic proteins of the Bcl-2 family initiate apoptosis by blocking the anti-apoptotic activity of Bcl-2 and Bcl-xL by binding to their mitochondrial binding sites or by triggering the activation of pro-apoptotic Bax/Bak.11 Cytochrome c is an intermediate in apoptosis, a controlled form of cell death used to kill cells in the process of development or in response to infection or DNA damage. Cytochrome c is released by the mitochondria in response to pro-apoptotic stimuli.12 Finding the exact molecular mechanism of apoptosis activation induced by maitake in breast cancer cells is our objective.
Evidence demonstrated that BAK-1 promotes cell death and counteracts the protection from apoptosis provided by Bcl-2.13,14 Chittenden et al.13 found that enforced expression of BAK-1 induced rapid and extensive apoptosis of serum-deprived fibroblasts, implying that BAK-1 may be directly involved in activating cell death machinery. Accordingly, Kiefer et al. reported that the BAK-1 gene product primarily enhances apoptotic cell death after an appropriate stimulus.14 In fact, during transduction of an apoptotic signal into the cell, an alteration in the permeability of the mitochondria membranes causes the translocation of the pro-apoptotic protein cytochrome c into the cytoplasm; this, in turn, activates death-driving proteolytic proteins known as caspases. Bcl-2 family members, which may play both anti-apoptotic or pro-apoptotic roles, regulate cell death by controlling this mitochondrial membrane permeability during apoptosis.
Our cDNA microarrays results showed that D fraction from maitake can induce the expression of 29 apoptosis-related genes; 22 of these are related to apoptosis induction and 7 are related to anti-apoptosis or apoptosis regulation. Positive and negative genetic and environmental regulators controls whether the pro-apoptotic or anti-apoptotic pathway is chosen. Pro-apoptotic gene activation will lead to cell death, while deactivation of the gene will block apoptotic pathways. Our results show that genetic regulation can also be modified by maitake treatment in MCF7 breast cancer cells.
Our study demonstrates that BAK-1 appears to be an essential gateway to the mitochondrial dysfunction that is required for cell death in response to maitake in MCF7 cells. However, more research must be done to determine the exact apoptosis activation and its regulation. We found that the pro-apoptotic messenger RNA VDAC-1 is significantly upregulated in the MCF7 cells treated with maitake in a dose-dependent manner; the increased reached 2.6-fold at 367μg of maitake per mL after 24 hours of treatment. We also found that maitake treatment upregulated Bcl-2-like 2 (13.21-fold increase with respect to control), a gene involve in the anti-apoptotic pathway. In 1999, Shimizu et al.15 showed that the recombinant pro-apoptotic proteins Bax (Bcl2-associated X protein) and BAK-1 accelerate the opening of VDAC, whereas the anti-apoptotic protein BCLXL closes VDAC by binding to it directly. Bax and BAK-1 allow cytochrome c to pass through VDAC out of liposomes, but passage is prevented by BCLXL. In agreement with this, VDAC-1-deficient mitochondria from mutant yeast did not exhibit a Bax/BAK-1-induced loss in membrane potential and cytochrome c release, both of which were inhibited by BCLXL. Shimizu et al.15 concluded that the Bcl-2 family of proteins binds to the VDAC in order to regulate the mitochondrial membrane potential and the release of cytochrome c during apoptosis. On the basis of this finding, we can postulate that maitake D fraction stimulates BAK-1 gene expression, thereby allowing cytochrome c to pass through VDAC-1 out of liposomes in MCF7 cells.
In this study, we found that VDAC-2 is upregulated by Maitake-treatment in MCF7 cells. Cheng et al. reported that BAK-1 aggregates with the mitochondrial outer membrane protein VDAC-2, a VDAC isoform present in low abundance that interacts specifically with the inactive conformer of BAK-1.16 Cells that are deficient in VDAC-2 but express the highly abundant VDAC-1 exhibited enhanced BAK-1 expression.16 Conversely, overexpression of VDAC-2 selectively prevented BAK-1 activation and inhibited the mitochondrial apoptotic pathway. From our results, we can probably assume that VDAC1 enhanced the expression of BAK-1 in the MCF7 cells after maitake treatment; however, the upregulation of VDAC-2 could partially block BAK-1 protein and prevent cell death by apoptosis in less that 40% of cells at 367μg of fraction D per mL, as was observed in the TUNEL assay results. Of note, this result shows that 60% of MCF7 cells treated with maitake will die as a result of apoptosis at the maximum concentration of D fraction after 24 hours of treatment.
BH3 interacting domain death agonist (BID) messenger RNA was found to be upregulated by maitake treatment in MCF7 cells (5.9-fold increase at 367μg/mL). Another pro-apoptotic activity occurs through the cytoplasmic protein, Bid. This molecule is found in the cytoplasm in an inactive form. When cleaved by activated caspase-8 from the extrinsic pathway, Bid causes a structural change to Bax, making it similar to the structure of the anti-apoptotic molecule, Bcl-2, and thereby allowing Bax to translocate to the mitochondria.17-20 Death signals activate "BH3-only" molecules, such as tBID, BIM, or BAD, which displace VDAC-2 from BAK-1, enabling homo-oligomerization of BAK-1 and apoptosis.16 Thus, we can conclude that VDAC-2 regulates the activity of BAK-1 and provides a connection between mitochondrial physiology and the core apoptotic pathway.
Many pro-apoptotic signals engage the apoptotic machinery by releasing cytochrome c from mitochondria.21-24 cDNA microarray data showed upregulation of apoptotic peptidase activating factor 1 interacting protein (APAF1) in breast cancer cells after maitake treatment in a dose-dependent manner. Cytoplasmic cytochrome c induces APAF-1 oligomerization, resulting in recruitment and activation of caspase 9 within the apoptosome.25-27 Caspase 9, in turn, is known to activate downstream effectors caspase 3 and 7.28 Finally, our cDNA microarray results demonstrate that caspase 7 and caspase 1 apoptosis-related genes in MCF7 cells were upregulated after 24 hours of maitake treatment (24.5- and 10-fold increases, respectively, at 183μg/mL; P<.001) (data not shown). It is well known that caspase proteins are the target key regulatory and structural proteins for proteolysis leading to cell death.28
β-glucans are not expressed on mammalian cells and are recognized as pathogen-associated molecular patterns by pattern recognition receptors, primarily the C-type lectin receptor dectin-1; they also interact via the complement receptor 3 (CR3).29-32 Dectin 1 is a small type II trans-membrane receptor with a lectin-like carbohydrate recognition domain that recognizes β1,3-linked and β1,6-linked glucans and intact yeast, while CR3 is a widely expressed β2 integrin containing a lectin domain, which mediates carbohydrate recognition.29-32 Maitake D fraction is rich in β1,3-linked glucans and probably interacts with a pattern recognition receptor in the cell membrane. A specific receptor for maitake in the mammalian cells has not yet been found.
On the basis of these results, Figure 6 summarizes the putative molecular mechanism involved in the apoptosis activation induced by G frondosa D fraction in breast cancer cells. Maitake triggers a cell stress signal pathway after interacting with the pattern recognition receptor in the cell membrane; it then induces the activation of the pro-apoptotic protein BAK-1 through activation of BID by caspase 1 or 7 and through activation of other BH3 initiator proteins. Activated BAK-1 proteins oligomerize at the mitochondrial membrane and cause the release of several mitochondrial factors, such as cytochrome c, which, in combination with APAF-1 and procaspase 7, form an apoptosome. Activated caspase 7 probably then activates caspase 1, allowing apoptosis to proceed. Bcl-2L2 blocks the activation of BAK-1 (Fig. 6).

FIG. 6. Putative molecular mechanism of apoptosis activation induced by maitake in MCF7 cells. This image summarizes the results obtained in this work. The gene expression involved in the apoptotic and anti-apoptotic pathway induced by D fraction in MCF7 cells is explained in the Discussion section. PRR, pattern recognition receptor.
In conclusion, the present study showed that the D fraction from maitake mushroom has strong anti-cancer properties in breast cancer cells, namely by exerting pro-apoptotic effects and reducing tumor cell viability. This action was accompanied by an increase in pro-apoptotic genes, which was further corroborated by an increase in BAK-1 gene expression, cytochrome c release to the cytoplasm, and activation of caspase 7 and caspase 1. Given the established epidemiologic and experimental findings regarding natural molecules, in particular from mushroom compounds, on cancer development and delay in progression, understanding the molecular mechanisms inherent in these effects is of utmost importance. This knowledge may lead to the development of therapeutic or preventive strategies for highly incident neoplasias, such as breast cancer.
ACKNOWLEDGMENTS
This study was partially funded by ERAB (EA-0641), Fundaçăo para a Ciência e Tecnologia (PTDC_EME_PME_70155_2006), and CONICET, Argentina.
REFERENCES
1. Parkin M: Global cancer statistics in the year 2000. Lancet Oncol 2001; 2: 533-543.
2. Hishida I, et al. Antitumor activity exhibited by orally administered extract from fruit body of Grifola frondosa (maitake). Chem Pharm Bull 1988; 36: 1819-1827.
3. Kodama N, Komuta K, Nanba H: Effect of maitake (Grifola frondosa) D-fraction on the activation of NK cells in cancer patients. Nutrition 2005; 21: 624-629.
4. Nanba H: Antitumor activity of orally administered D-fraction from maitake mushroom. J Naturopathic Med 1993; 1: 10-15.
5. Ohno N, et al: Structural characterization and antitumor activity of the extracts from matted mycelium of cultured Grifola frondosa. Chem Pharm Bull 1985; 33: 3395-3401.
6. Soares R, Guerreiro S, Botelho M: Elucidating progesterone effects in breast cancer: cross talk with PDGF signalling pathway in smooth muscle cells. J Cell Biochem 2007; 100: 174-183.
7. Greenie RT, Murray T, Boldin S, Wingo P: Cancer statistics 2000. CA Cancer J Clin 2000; 50: 7-23.
8. Stewart BW, Kleihues P, eds. World Cancer Report. IARC Press, Lyon, France, 2003.
9. Ramljak D, Romanczyk LJ, Metheny-Barlow L, et al: Pentameric procyanidin from Theobroma cacao selectively inhibits growth of human breast cancer cells. Mol Cancer Therap 2005; 4: 537-546.
10. Kozikowski AP, Tuckmantel W, Bottcher G, Romanczyk LJ Jr: Studies in polyphenol chemistry and bioactivity and their inhibition of cancer cell growth through cell cycle arrest. J Organic Chem 2003; 68: 1641-1658.
11. Shomori K, Yamamoto M, Arifuku I, Teramachi K, Ito H: Antitumor effects of a water-soluble extract from Maitake (Grifola frondosa) on human gastric cancer cell lines. Oncol Rep 2009; 22 :615-620.
12. Liu X, Kim C, Yang J, Jemmerson R, Wang X: Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 1996; 86: 147-157.
13. Chittenden T., Harrington EA., O’Connor R., et al: Induction of apoptosis by the Bcl-2 homologue BAK-1. Nature 1995; 374: 733-736.
14. Kiefer MC, Brauer MJ, Powers VC., et al: Modulation of apoptosis by the widely distributed Bcl-2 homologue BAK-1. Nature 1995; 374: 736-739.
15. Shimizu S., Narita M, Tsujimoto Y: Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature 1999; 399: 483-487.
16. Cheng EHY, Sheiko TV, Fisher JK, et al: VDAC2 inhibits BAK-1 activation and mitochondrial apoptosis. Science 2003;301: 513–517.
17. Luo X, Budihardjo I, Zou H, et al: Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 1998; 94: 481-490.
18. Li H, Zhu H, Xu CJ, et al: Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 1998;94:491-501.
19. Gross A, Yin XM, Wang K, et al: Caspase cleaved BID targets mitochondria and is required for cytochrome c release, while BCL-XL prevents this release but not tumor necrosis factor- R1/ Fas death. J Biol Chem 1999; 274: 1156-1163.
20. Ruffolo SC, Breckenridge DG, Nguyen M, et al: BID-dependent and BID-independent pathways for BAX insertion into mitochondria. Cell Death Differ 2000; 7: 1101-1108.
21. Allan LA, Clarke PR: Apoptosis and autophagy: regulation of caspase-9 by phosphorylation. FEBS J. 2009; 276: 6063-6073.
22. Liu X, Kim CN, Yang J, Jemmerson R, Wang X: Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 1996;86:147-157.
23. Kluck RM, Bossy-Wetzel E, Green DR, Newmeyer DD: The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science 1997; 275: 1132-1136.
24. Yang Y, Liu X, Bhalla K, et al: Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science 1997; 275: 1129-1132.
25. Li P, Nijhawan D, Budihardjo I, et al: Cytochrome c and dATP-dependent formation of APAF-1/ caspase-9 complex initiates an apoptotic protease cascade. Cell 1997; 91: 479-489.
26. Zou H, Henzel WJ, Liu X, Lutschg A, Wang X: Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 1997; 90: 405-413.
27. Riedl SJ, Salvesen GS: The apoptosome: signaling platform of cell death. Nat Rev Mol Cell Biol 2007; 8: 405-413.
28. Taylor RC, Cullen SP, Martin SJ: Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol 2008; 9: 231-241.
29. Brown GD, Gordon S. Immune recognition. A new receptor for beta-glucans. Nature 2001; 413: 36-37.
30. Brown GD, Herre J, Williams DL, Willment JA, Marshall AS, Gordon S: Dectin-1 mediates the biological effects of beta-glucans. J Exp Med 2003; 197: 1119-1124.
31. Willment JA, Gordon S, Brown GD. Characterization of the human beta-glucan receptor and its alternatively spliced iso-forms. J Biol Chem 2001; 276: 43818-43823.
32. Netea MG, Brown GD, Kullberg BJ, Gow NA: An integrated model of the recognition of Candida albicans by the innate immune system. Nat Rev Microbiol 2008; 6; 67-78.
本文部分内容来源网络,如有侵权请联系删除,谢谢。
本文部分内容来源网络,如有侵权请联系删除,谢谢。
编辑:中润世源
上一篇:荷瘤小鼠口服舞茸中可溶性β-葡聚糖能诱导全身 下一篇:没有了
